Monitor netCDF File - Sample Program
This Fortran
progam creates a netCDF file containing 3 variables: r, time,
and
density. r is a 1-dimensional variable
containing six X values of points to plot. density is a
2-dimensional variable containing 11 sets of six Y values of points to
plot. time is a
1-dimensional variable
with 11 values that are used as time indices to the sets of Y
values. The program runs through 11 time steps. In each
time step it adds to the time and density variables. They are
defined with an "unlimited" dimension within the file so they are
extended at each time step. At time step 7, e.g., time contains only 7 values and density contains only 7 sets.
ElVis tracks the size of variables and updates the graph when their
size changes. Define
variables to be monitored with an unlimited
dimension so ElVis can detect a change in their size.
The attribute named "running" is set to true until the final time
step when it is set to false. ElVis will monitor the file while
running is set to true.
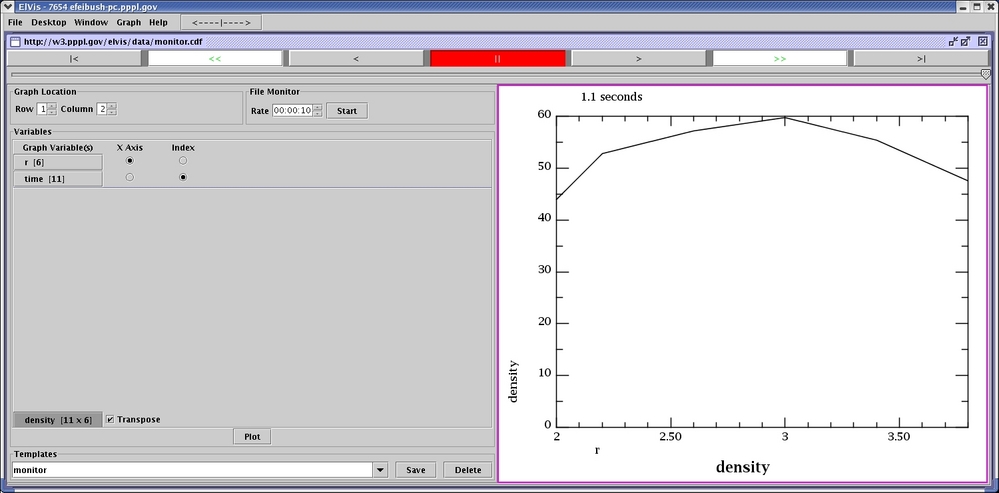
The output file is in
http:/w3.pppl.gov/elvis/data/monitor.cdf. It has a corresponding
template file, monitor.cdft. When you run ElVis and browse
for monitor.cdf the resulting plot is shown below the sample
program. The netCDF panel lists the variables in the file. r is selected as the X-axis values
and time is selected as the
index. The density
variable is selected for plotting. Its data is transposed
because array indexing in ElVis (Java) is transposed from
Fortran. The graph is an animated f(x,i) showing, in this screen
shot, the final time step. The slider or the VCR buttons in ElVis
are used to change the index to a different time step.
Fortran 90 netCDF Interface
program simmonitor
use netcdf
implicit none
C netCDF file ID
integer ncid
C netCDF error status return
integer iret
C netCDF dimension IDs
integer samples_dim
integer timesteps_dim
C program variables for dimensions
integer samples_len
parameter (samples_len = 6)
integer timesteps_len
parameter (timesteps_len = 11)
integer start(1)
integer start_den(2)
integer count(1)
integer count_den(2)
C netCDF variable IDs
integer time_id
integer r_id
integer density_id
C netCDF variable shapes
integer time_dims(1)
integer r_dims(1)
integer density_dims(2)
C program data variables
real*8 r(samples_len)
data r / 2., 2.4, 2.8, 3.2, 3.6, 4. /
real*8 simtime(1)
real*8 density(samples_len)
integer i, j
real*8 densitycalc
C create netCDF file and enter
define mode
iret = nf90_create('monitor.cdf', NF90_CLOBBER, ncid)
call check_err(iret)
C define netCDF dimensions
iret = nf90_def_dim(ncid, 'samples', samples_len, samples_dim)
call check_err(iret)
iret = nf90_def_dim(ncid, 'timesteps', NF90_UNLIMITED,
> timesteps_dim)
call check_err(iret)
C define netCDF variables
r_dims(1) = samples_dim
iret = nf90_def_var(ncid, 'r', NF90_DOUBLE, r_dims, r_id)
call check_err(iret)
time_dims(1) = timesteps_dim
iret = nf90_def_var(ncid, 'time', NF90_DOUBLE, time_dims,
>
time_id)
call check_err(iret)
iret = nf90_put_att(ncid, time_id, 'units', 'seconds')
call check_err(iret)
density_dims(1) = samples_dim
density_dims(2) = timesteps_dim
iret = nf90_def_var(ncid, 'density', NF90_DOUBLE,
>
density_dims, density_id)
call check_err(iret)
C set netCDF attribute for ElVis
to start monitoring netCDF file
iret = nf90_put_att(ncid, NF90_GLOBAL, 'running', 'true')
call check_err(iret)
C leave netCDF define mode
iret = nf90_enddef(ncid)
call check_err(iret)
C put data into netCDF r variable
iret = nf90_put_var(ncid, r_id, r)
call check_err(iret)
iret = nf90_sync(ncid)
call check_err(iret)
C initialize the simulation
simtime(1) = 0.
count(1) = 1
count_den(1) = samples_len
count_den(2) = 1
start_den(1) = 1
C loop through the simulation
time steps
do
j=1,timesteps_len
simtime(1) = simtime(1) + .1
C append the
current time value to the 1-D time variable in the file
start(1) = j
iret = nf90_put_var(ncid, time_id, simtime, start, count)
call check_err(iret)
C calculate
density for this time step
do i=1,samples_len
density(i) = densitycalc(i, j)
enddo
C append the
1-D density to the 2-D density variable in the file
start_den(2) = j
iret = nf90_put_var(ncid, density_id, density, start_den,
> count_den)
call check_err(iret)
iret = nf90_sync(ncid)
call check_err(iret)
print *, j
call sleep(5)
enddo
C set netCDF attribute for ElVis
to stop monitoring netCDF file
iret = nf90_redef(ncid)
call check_err(iret)
iret = nf90_put_att(ncid, NF90_GLOBAL, 'running', 'false')
call check_err(iret)
iret = nf90_enddef(ncid)
call check_err(iret)
iret = nf90_close(ncid)
call check_err(iret)
end
real function densitycalc(i, step)
implicit none
integer i
integer step
real initd(6)
data initd / 5., 6., 6.5, 6.8, 6.3, 5.4 /
real dcalc
if
(step .eq. 1) then
dcalc = initd(i)
else
dcalc = initd(i) * step * .8
endif
densitycalc = dcalc
return
end
subroutine check_err(iret)
use netcdf
implicit none
integer iret
if
(iret .ne. nf90_NoErr) then
print *, nf90_strerror(iret)
stop
endif
end
Fortran 77 netCDF Interface
program simmonitor
implicit none
include
'netcdf.inc'
C netCDF file ID
integer ncid
C dimension IDs
integer samples_dim
integer timesteps_dim
C dimensions
integer samples_len
integer timesteps_len
parameter (samples_len = 6)
parameter (timesteps_len = 11)
integer start(1)
integer count(1)
C variable IDs
integer time_id
integer r_id
integer density_id
C variable shapes
integer time_dims(1)
integer r_dims(1)
integer density_dims(2)
C variable data
real time(timesteps_len)
real r(samples_len)
real density(samples_len, timesteps_len)
C error status return
integer iret
integer i, j
real simtime
real densitycalc
C create netCDF file and enter
define mode
iret = nf_create('monitor.cdf', NF_CLOBBER, ncid)
call check_err(iret)
C define dimensions
iret = nf_def_dim(ncid, 'samples', 6, samples_dim)
call check_err(iret)
iret = nf_def_dim(ncid, 'timesteps', NF_UNLIMITED, timesteps_dim)
call check_err(iret)
C define variables
r_dims(1) = samples_dim
iret = nf_def_var(ncid, 'r', NF_REAL, 1, r_dims, r_id)
call check_err(iret)
time_dims(1) = timesteps_dim
iret = nf_def_var(ncid, 'time', NF_REAL, 1, time_dims,
1
time_id)
call check_err(iret)
iret = nf_put_att_text(ncid, time_id, 'units', 7, 'seconds')
call check_err(iret)
density_dims(1) = samples_dim
density_dims(2) = timesteps_dim
iret = nf_def_var(ncid, 'density', NF_REAL, 2,
1
density_dims, density_id)
call check_err(iret)
C set attribute for ElVis to
start monitoring netCDF file
iret = nf_put_att_text(ncid, NF_GLOBAL, 'running', 4, 'true')
call check_err(iret)
C leave define mode
iret = nf_enddef(ncid)
call check_err(iret)
C put data into r variable
data r / 2., 2.4, 2.8, 3.2, 3.6, 4. /
iret = nf_put_var_real(ncid, r_id, r)
call check_err(iret)
iret = nf_close(ncid)
call check_err(iret)
C initialize the simulation
simtime = 0.
start(1) = 1
C run the simulation
do
j=1,timesteps_len
simtime = simtime + .1
iret = nf_open('monitor.cdf', NF_WRITE, ncid)
call check_err(iret)
time(j) = simtime
count(1) = j
iret = nf_put_vara_real(ncid, time_id, start, count, time)
call check_err(iret)
do i=1,6
density(i,j) = densitycalc(i, j)
enddo
iret = nf_put_var_real(ncid, density_id, density)
call check_err(iret)
iret = nf_close(ncid)
call check_err(iret)
enddo
C assign attribute for ElVis to
stop monitoring output file
iret = nf_open('monitor.cdf', NF_WRITE, ncid)
call check_err(iret)
iret = nf_redef(ncid)
call check_err(iret)
iret = nf_put_att_text(ncid, NF_GLOBAL, 'running', 5, 'false')
call check_err(iret)
iret = nf_enddef(ncid)
call check_err(iret)
iret = nf_close(ncid)
call check_err(iret)
end
real function densitycalc(i, step)
implicit none
integer i
integer step
real initd(6)
data initd / 5., 6., 6.5, 6.8, 6.3, 5.4 /
real dcalc
if
(step .eq. 0) then
dcalc = initd(i)
else
dcalc = initd(i) * step * .8
endif
densitycalc = dcalc
return
end
subroutine check_err(iret)
implicit none
integer iret
include 'netcdf.inc'
if
(iret .ne. NF_NOERR) then
print *, nf_strerror(iret)
stop
endif
end